In scientific research, SNP is a term that is frequently used, but what is SNP exactly, why do we study SNP, and how do we carry out research work?
What is SNP?
SNP, or Single nucleotide polymorphism, refers to the diversity of DNA sequences attributable to insertions or deletions, single nucleotide transitions, and transversions with a variation frequency of >1%.
SNPs are abundant, and there are around 3 × 106 SNP sites in the human body, with an average of 1 in 500-1000 base pairs. Among the vast number of SNP loci in the human body, it is limited to those that can cause amino acid changes, contingent on the position of the SNP and the type of mutation.
As illustrated in the figure below, only SNPs with non-synonymous mutations in the coding gene region produce phenotypic changes.
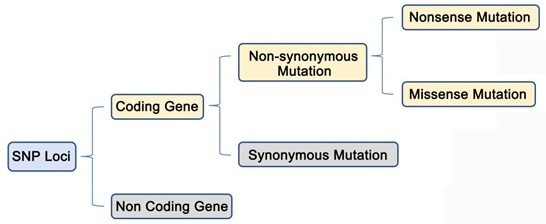
Image Credit: Nanjing Vazyme Biotech Co., Ltd
SNPs are affiliated with drug susceptibility, disease susceptibility and individual phenotypic differences. They possess significant research value in precise nutrition, disease diagnosis and screening, and medication guidance. They are closely connected to our daily lives.
Studies have shown that SNP loci and related genes which may be associated with the symptoms of COVID-19 have been discovered.1 During its evolution, numerous SNP loci appeared in the COVID-19 virus, some of which could create a new variant of the coronavirus disease that has a potential risk of increasing virulence and infectiousness.
Research on SNPs can be split into two categories:
- Evaluation of unknown SNPs, such as identifying new SNP sites and establishing the relationship between an unknown SNP and genetic disease.
- Analysis of known SNPs, such as genetic diversity studies of SNPs across various groups and genetic diagnosis of genetic diseases.
The most commonly used methods for detecting SNPs
SNP detection is primarily carried out by PCR and sequencing. The base and site of mutation can be established using detection. These methods, according to the detection throughput, can be split into two main categories: low-throughput and high-throughput.
The low-throughput method has the potential to detect dozens of SNPs in one experiment, while the high-throughput method can detect SNPs in their thousands at one time.
Low-throughput methods include Sanger sequencing, Taqman probes, and mass spectrometry detection.
Sanger sequencing
Sanger sequencing relies on dideoxynucleotide termination reaction to produce fragments that vary in length for sequencing. Considered the “gold standard” for SNP detection, Sanger sequencing not only has the capacity to determine the type and location of mutations but also identify unknown SNP sites.
The throughput of Sanger sequencing is low, and the cost is comparatively high. Sanger sequencing is appropriate for analyzing fewer sites and fewer samples.

